Vocabulary Normalization
Vocabulary normalization is the process of standardizing the codes that make up healthcare data. Healthcare data is made up of millions of atomic-level codes from a number of coding systems (e.g. SNOMED-CT, RxNorm, LOINC, and ICD-10). These codes are frequently missing from raw healthcare data sources because the source systems that generate the data are not setup properly. As a result, raw data sources may include descriptions (e.g. Fasting Glucose) but not have the associated standard codes (e.g. the LOINC for Fasting Glucose). Because nearly all downstream analytics and research use cases depend on these standard codes, this severely limits the utility of your data.
This problem is more common in clinical data sources (e.g. EHR data) than claims data. One of the good things about claim data is that the codes are fairly standardized. This is because claims are used for billing and a claim will be denied if it doesn't have the appropriate codes.
We've engineered Tuva to allow you to map non-standard codes and descriptions to standard codes. This process is generally referred to as vocabularly normalization or terminology normalization. In the sections that follow we describe how you can use the Tuva pipelines to insert standard codes. In the current state you must generate the mappings, but Tuva gives you a straightforward way to insert in these mappings into the data model. We are in the process of developing a normalization engine that will automatically create the mappings.
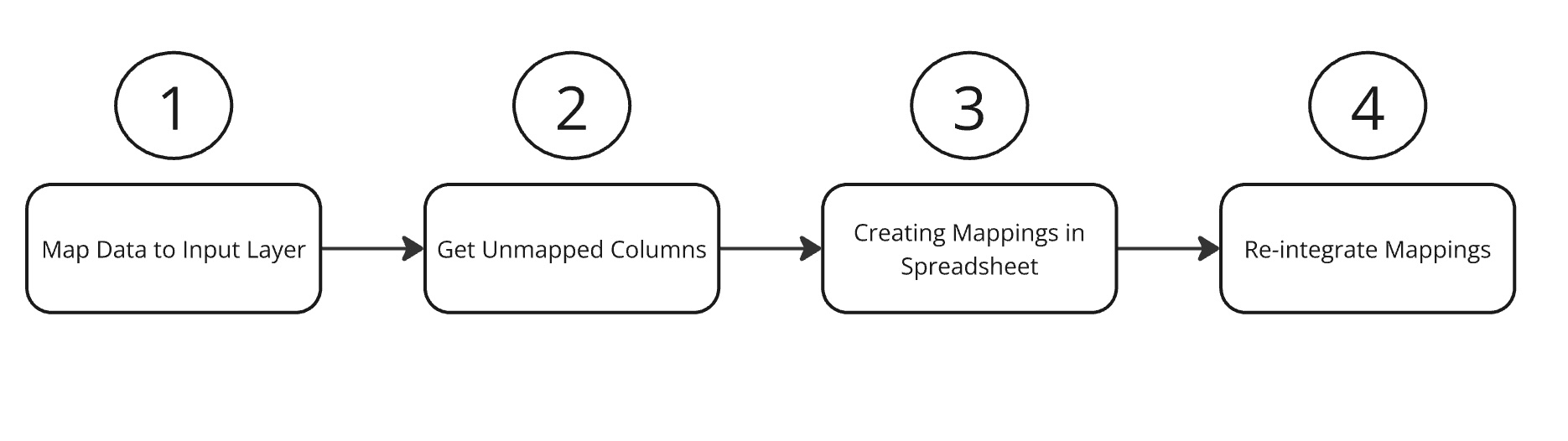
1. Process Overview
The Input Layer and Core Data Model tables have two sets of columns to define the code associated with each record: the source columns, and the normalized columns. The condition, procedure, lab_result, immunization and observation tables all have source_code_type, source_code, and source_description columns, as well as normalized_code_type, normalized_code, and normalized_description columns. The medication table has the same source columns, and
separate normalized columns for ndc_code and ndc_description, rxnorm_code and rxnorm_description, and atc_code and atc_description.
The intent is to populate the source columns with the values present in the source system, and to populate the normalized columns with standardized terminology code sets. The Data Marts will look first in the normalized columns before the source columns when looking for qualifying records for analysis (e.g. for running quality measures).
Out of the box, if the normalized fields are left null in the Input Layer, Tuva will try to populate them. Data pipelines in the Core Data Model layer will check if the source code value is a standardized terminology code type, i.e. icd-10-cm, icd-9-cm, icd-10-pcs, icd-9-pcs, hcpcs, snomed-ct, loinc, ndc, cvx or rxnorm depending on the model. If the code type is one of these standard code types, it will compare the source_code to the relevant Tuva terminology dictionary, and if it finds a valid match it will populate the normalized fields.
This works slightly differently in the Medication table in the Core Data Model layer. For medications if the source code type is an NDC or RxNorm, Tuva will also try to populate RxNorm and ATC level 3 codes.
If a user populates the normalized columns in their Input Layer models, Tuva will respect those values regardless of if they are valid, and persist those values through to the Core Data Model. Each table in the Core Data Model also has a mapping_method column that will be manual if the value was populated by the user in the input layer, or automatic if the value was populated by the tuva project.`
2. Getting Unmapped Codes
Tuva has a built-in process for integrating custom mappings to standardized terminologies, which can be configured through an optional enable_normalize_engine var in Tuva.
The first step is to produce a list of unmapped codes. Setting enable_normalize_engine: unmapped in the dbt_project.yml will enable a new normalize mart Tuva. This mart will initially contain an all_unmapped table that has all of the unmapped codes across all domains, as well as individual unmapped_condition,unmapped_procedure, unmapped_medication, unmapped_lab_result, unmapped_immunization, and unmapped_observations tables. These tables will contain a list of codes that weren't able to be automatically mapped and weren't manually mapped to normalized codes, as well as counts and a list of domains the codes appear in, and other columns to support the mapping process.
Note that one source of false positives is HCPCS Level 1 or CPT codes. Due to licensing restrictions from the AMA, Tuva isn't able to include a dictionary to validate CPT codes, so HCPCS codes will only be evaluated against a HCPCS level 2 dictionary.
3. Creating Code Mappings
The next step is to create mappings for the unmapped codes. The all_unmapped table can be exported and used as a mapping workbook; it contains all of the columns that Tuva needs to reintegrate the maps into the Core Data Model.
For a given row, if a code is mappable to a standardized terminology, a user should populate normalized_code_type,normalized_code, and normalized_description with the normalized values, and Tuva will populate those values when matching on the source_code_type, source_code, and source_code_description values in that row. Alternatively, if the code is not mappable to a standardized terminology, the user can populate a reason in the not_mapped. If either not_mapped or the normalized fields are populated when the workbook is reintegrated into the tuva project, the codes won't be in the unmapped table in subsequent runs.
The all_unmapped table also contains additional columns to facilitate the mapping process. It has added_by and added_date columns to record who created the map and when, reviewed_by and reviewed_date to record the reviewer, as well as a notes columns to record any extra information the mapper would like to record about the mapping. These columns will be persisted in the Tuva data model, but will not be present in the Core Data Model tables.
The standard workflow we follow for creating code mappings works something like this.
- Export the
all_unmappedtable and copying it into a spreadsheet. - Sort the spreadsheet by term frequency and the manually start mapping the correct codes. This often requires clinical informatics subject matter expertise.
- Once mapping is complete have a second person review the mappings for accuracy.
- Once the mappings are ready to be re-integrated we follow the steps below.
4. Re-integrating Code Mappings
To re-integrate the code mappings, the user needs to add a model or seed in their dbt project called custom_mapped that contains the data from the mapping workbook. The user can choose to keep the entire workbook in the project as a seed, to keep an empty seed with only the required headers and populate the table from a cloud storage service with our load_seed() macro, or they can maintain the mappings in their data warehouse and have custom_mapped be a model that selects the required columns.
Once the user has custom_mapped added to their project, they can set the enable_normalize_engine to true, and on the subsequent run, the Tuva will integrate the normalized codes from the mapping workbook into the normalized colums of the core tables. In addition, the normalized mart will now also have an all_codes model, that contains all of the existing custom_mapped codes as well as any codes that are unmapped, so that table can be exported to build a new complete mapping workbook if desired. Any codes that are mapped with custom_mapped will show custom in their corresponding Core Data Model tables' mapping_method columns.